

- #Microarray probe to gene r expression taply how to
- #Microarray probe to gene r expression taply software
Quantile normalization was then performed and inter-array correlations were calculated using R.
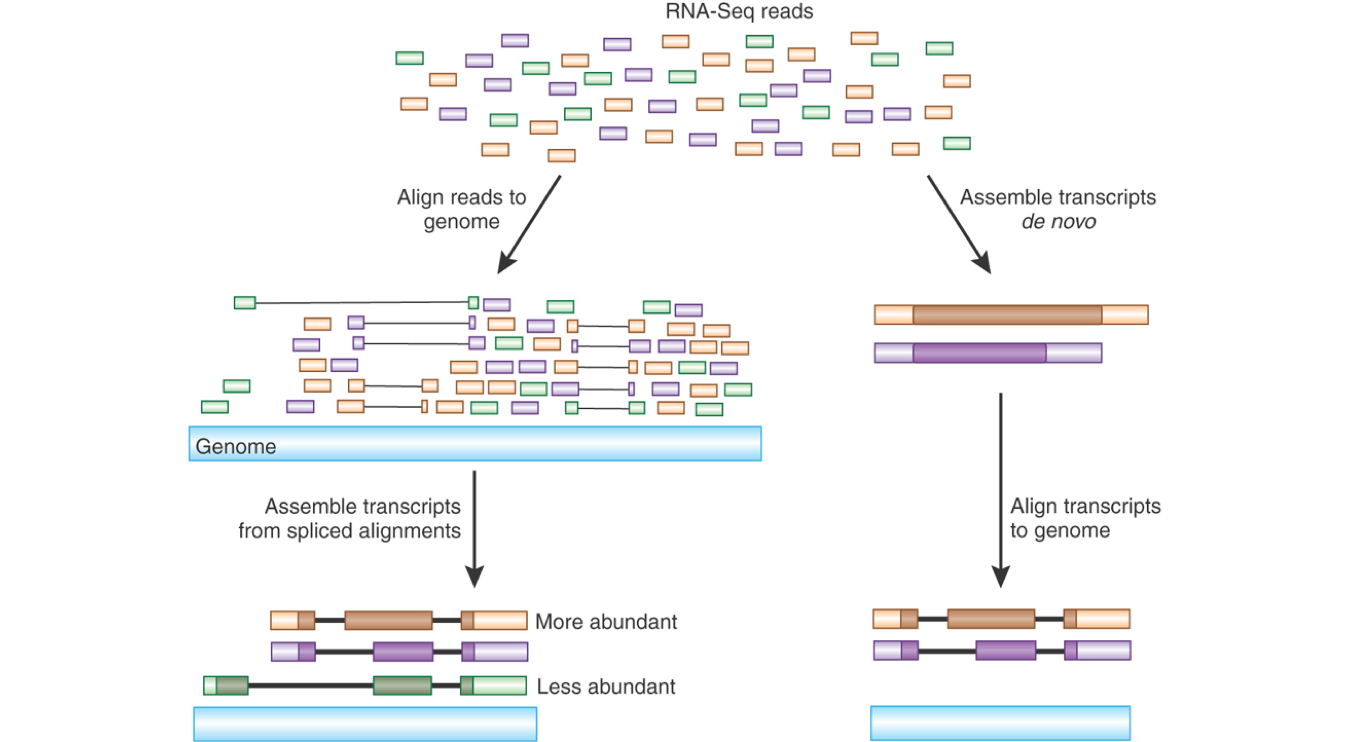
For each array, expression values were scaled to an average intensity of 200 (GCOSv1.2, Affymetrix, Inc.). In addition, only probe sets with six or more matching probes were retained for subsequent analyses (n=11,768/12,625). Any probe without a perfect match in both species (approximately 1/4) was masked during the calculation of expression values (GCOSv1.2, Affymetrix, Inc). All probes on the array were compared against the human genome (Build 34) and the chimpanzee draft genome using MegaBlast (). Because the arrays used in this study were designed for human mRNA sequences, it is necessary to account for the effect of interspecies sequence differences on chimpanzee gene-expression values. The dataset used for network construction consisted of 36 Affymetrix HGU95Av2 microarrays surveying gene expression with 12,625 probe sets in three adult humans and three adult chimpanzees across six matched brain regions: Broca's area, anterior cingulate cortex, primary visual cortex, prefrontal cortex, caudate nucleus, and cerebellar vermis. Analysis of microarray data The following description is taken from Oldham et al 2006. To facilitate comparison with the original analysis, we use the microarray normalization procedures and gene selection procedures described in Oldham et al (2006). Nov 21 103(47):17973-8 This tutorial and the data files can be found at the webpage More material on weighted network analysis can be found at The unprocessed data came originally from Khaitovich et al (2004). BMC Bioinformatics The microarray data and processing steps are described in Oldham M, Horvath S, Geschwind D (2006) Conservation and Evolution of Gene Coexpression Networks in Human and Chimpanzee Brains. The methods and biological implications are described in the following reference Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules.
#Microarray probe to gene r expression taply software
Some familiarity with the R software is desirable but the document is fairly selfcontained. The R code allows to reproduce the Figures and tables reported in Langfelder and Horvath (2007). The two data sets correspond to gene expression measurements in human and chimpanzee brains.
#Microarray probe to gene r expression taply how to
Eigengene Network Analysis of Human and Chimpanzee Microarray Data R Tutorial Peter Langfelder and Steve Horvath Correspondence:, This is a self-contained R software tutorial that illustrates how to carry out an eigengene network analysis across two datasets.


 0 kommentar(er)
0 kommentar(er)
